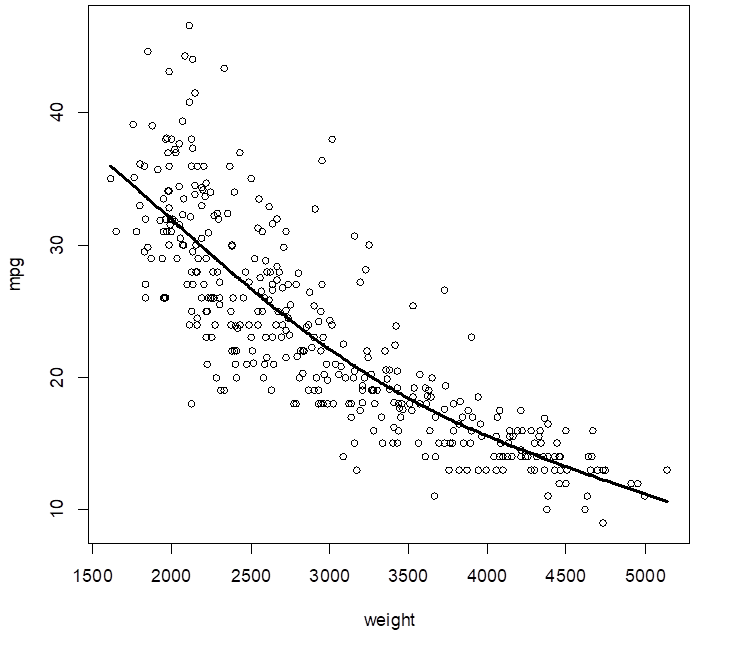#
Dr. M. Baron, Statistical Machine Learning class, STAT-427/627
# POLYNOMIALS AND SPLINES
1. Fitting a polynomial regression
> attach(Auto);
> plot( weight, mpg )
> reg = lm( mpg ~ weight )
> abline(reg)
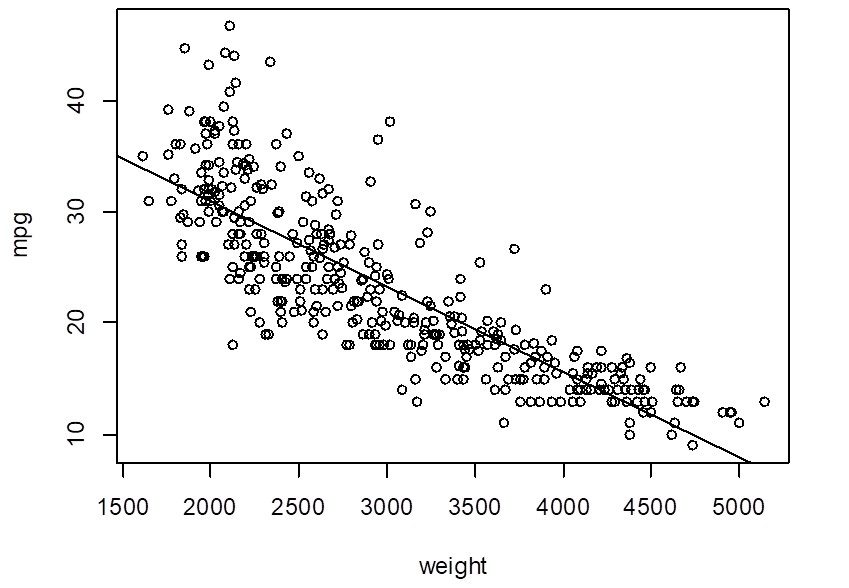
# Look at the residuals. Is linear model adequate?
> plot( weight, residuals(reg) )
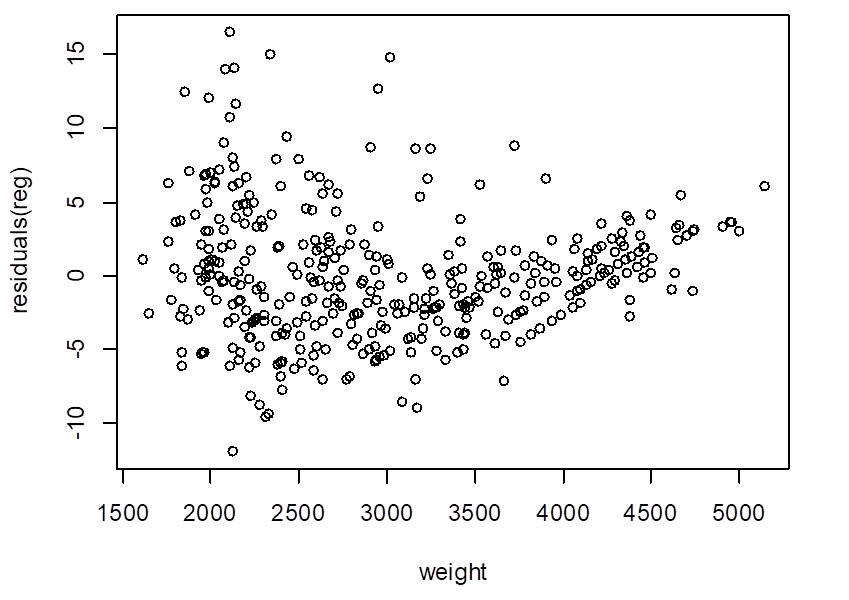
# If a polynomial regression is appropriate,
what is the best degree?
>
install.packages("leaps")
> library(leaps)
> polynomial.fit = regsubsets(
mpg ~ poly(weight,10), data=Auto )
> summary(polynomial.fit)
poly(weight, 10)1
poly(weight, 10)2 poly(weight, 10)3 poly(weight, 10)4 poly(weight, 10)5
poly(weight, 10)6
1 ( 1 ) "*" " " " " " " " " " "
2 ( 1 ) "*" "*" " " " " " " " "
3 ( 1 ) "*" "*" " " " " " " " "
4 ( 1 ) "*" "*" " " " " " " " "
5 ( 1 ) "*" "*" " " " " "*" " "
6 ( 1 ) "*" "*" " " " " "*" "*"
7 ( 1 ) "*" "*" " " " " "*" "*"
8 ( 1 ) "*" "*" " " "*" "*" "*"
poly(weight, 10)7 poly(weight, 10)8
poly(weight, 10)9 poly(weight, 10)10
1 ( 1 ) " " " " " " " "
2 ( 1 ) " " " " " " " "
3 ( 1 ) " " " " " " "*"
4 ( 1 ) "*" " " " " "*"
5 ( 1 ) "*" " " " " "*"
6 ( 1 ) "*" " " " " "*"
7 ( 1 ) "*" " " "*" "*"
8 ( 1 ) "*" " " "*" "*"
> which.max(summary(polynomial.fit)$adjr2)
[1] 4
> which.min(summary(polynomial.fit)$cp)
[1] 3
> which.min(summary(polynomial.fit)$bic)
[1] 2
# BIC chooses a quadratic model “mpg
= β0 + β1 (weight) + β2(weight)2
+ ε”. Mallows Cp and adjusted R2 add higher order terms.
# Cross-validation agrees with the quadratic model…
> library(boot)
> cv.error =
rep(0,10)
> for (p in 1:10){
+ polynomial = glm( mpg ~ poly(weight,p) )
+ cv.error[p] = cv.glm(
Auto, polynomial )$delta[1] }
> cv.error
[1]
18.85161 17.52474 17.57811 17.62324 17.62822 17.69418 17.66695 17.76456
18.35543 18.59401
> plot(cv.error);
lines(cv.error)
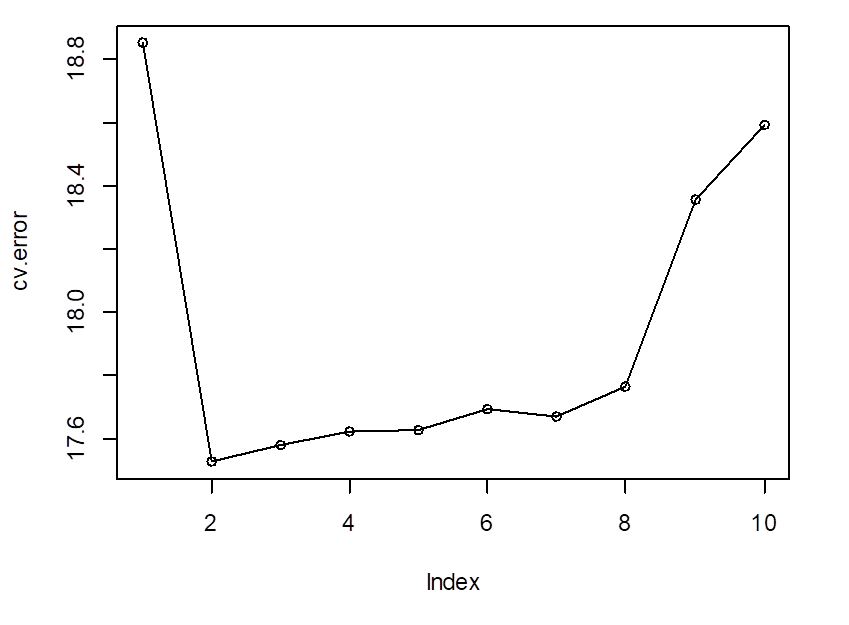
# So, we choose the quadratic regression – degree 2 polynomial. Its prediction MSE is
17.52.
> poly2 = lm( mpg ~
poly(weight,2) )
> summary(poly2)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 23.4459 0.2109 111.151 < 2e-16 ***
poly(weight,
2)1 -128.4436 4.1763 -30.755 < 2e-16 ***
poly(weight,
2)2 23.1589 4.1763
5.545 5.43e-08 ***
> plot( weight,mpg
)
> Yhat_poly2 = fitted.values(
poly2 )
> points( weight, Yhat_poly2,
col="red", lwd=3 )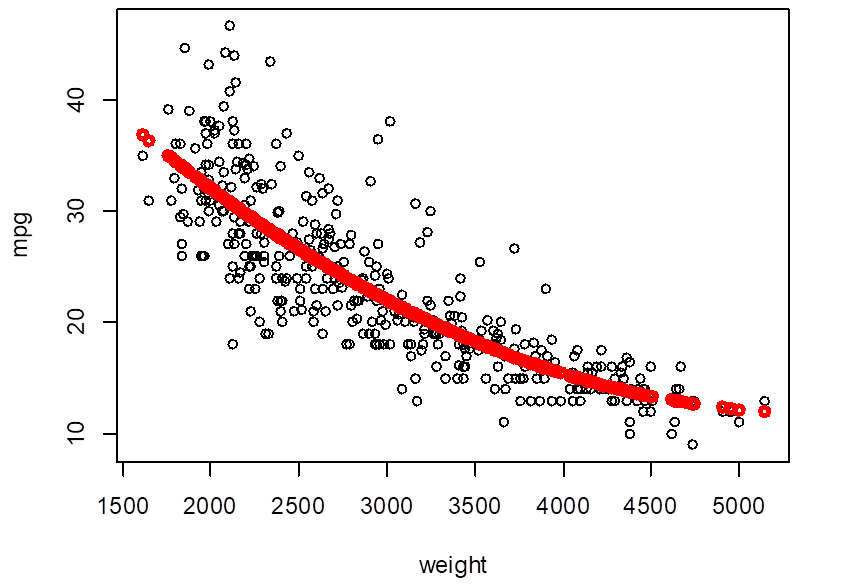
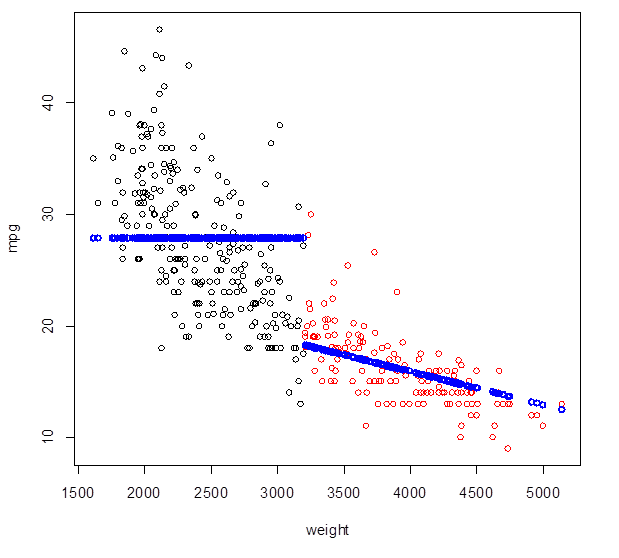
2. Broken Line – fitting different models on different intervals of X.
> n = length(mpg); # From the residual plot, the trend
> Group = 1*(weight > 3200) # changes around weight = 3200 lbs
> plot(weight, mpg, col = Group+1 ) # Red points correspond to heavier cars
> broken_line = lm( mpg ~ weight : Group ) # Interactions allow different slopes
> Yhat_broken =
predict(broken_line) # Fitted values
> points( weight, Yhat_broken,
lwd=2, col="blue" ) # Add fitted
values to the plot
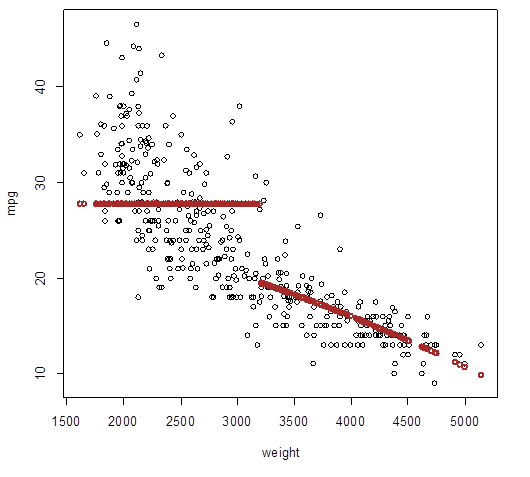
# Quadratic polynomial fit does not improve the picture
> broken_quadratic
= lm( mpg ~ (weight + I(weight^2)) : Group )
> Yhat_broken_quadratic
= predict(broken_quadratic)
> plot(weight,mpg); points( weight, Yhat_broken,
lwd=2, col="brown" )
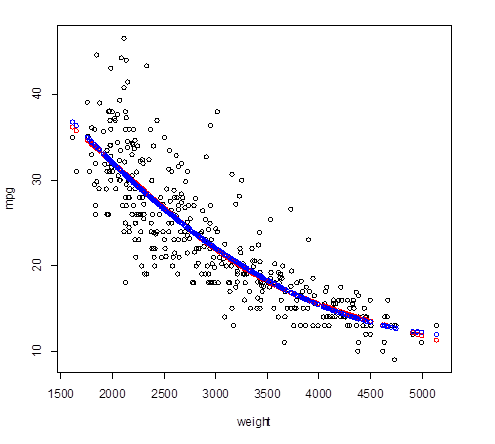
3. Splines – smooth connection at
knots.
> install.packages("splines")
>
library(splines)
> spline = lm( mpg
~ bs(weight, knots=3200)
) # In this lm, we
defined the basis and knots
> Yhat_spline =
predict(spline) # Fitted values
> plot(weight,mpg); points( weight, Yhat_spline,
col="red" )

# This spline is almost the same as the quadratic polynomial
regression!
> points( weight, Yhat_poly2,
col="blue" )
# Add more knots?
> spline = lm( mpg
~ bs(weight, knots=c(2000,2500,3200,4500)) )
> Yhat_spline = predict(spline)
> plot(weight,mpg); points( weight, Yhat_spline,
col="red" )
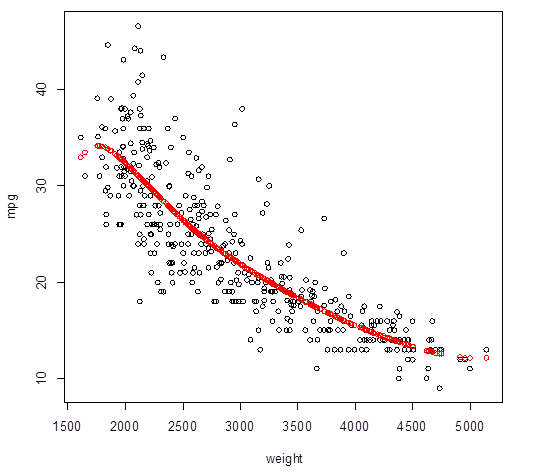
4. Cross-validation
> library(boot)
# Compare these four models by K-fold cross-validation, with K=n/4
groups, deleting 4 units at a time.
> reg = glm( mpg ~ weight
)
> poly2 = glm( mpg
~ poly(weight,2) )
> spline1knot = glm(
mpg ~ bs(weight, knots=3200) )
> spline4knots = glm(
mpg ~ bs(weight, knots=c(2000,2500,3200,4500)) )
> cv.glm( Auto,
reg, K=n/4 )$delta[1]
[1] 18.84014
> cv.glm( Auto,
poly2, K=n/4 )$delta[1]
[1] 17.51702
> cv.glm( Auto,
spline1knot, K=n/4 )$delta[1]
[1] 17.59429
> cv.glm( Auto,
spline4knots, K=n/4 )$delta[1]
[1] 17.81641
# The quadratic polynomial model wins. Splitting X range into
segments for the splints did not make much difference.
5. Smoothing Splines
5a. Fitting a smoothing spline
> attach(Auto); plot(weight,mpg); attach(splines);
> ss5 = smooth.spline(weight, mpg, df=5)
> lines(ss5, col="red", lwd=3)
> ss25 = smooth.spline(weight,
mpg, df=25)
> lines(ss25, col="blue", lwd=3)
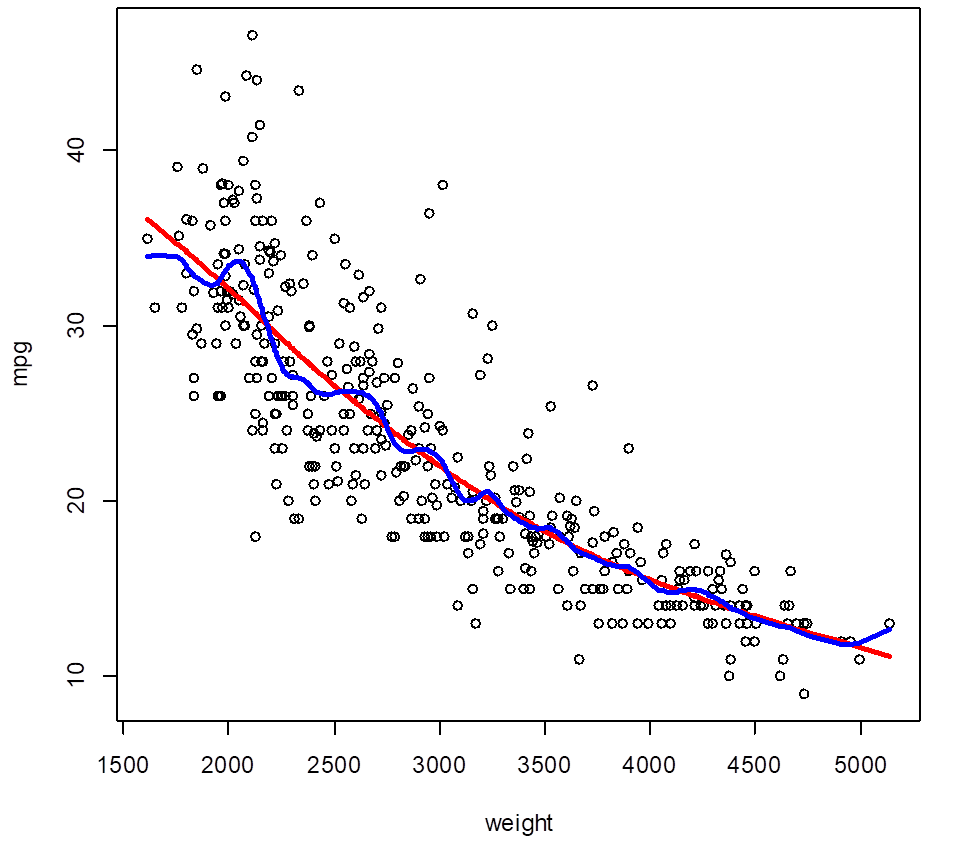
# A spline with (an equivalent of) 5 degrees of freedom is very
smooth, its flexibility is limited by small df. A
spline with 25 df is probably too flexible, too
wiggly. The optimal df can be determined by
cross-validation.
5b. Prediction error and cross-validation
> n = length(mpg); Z =
sample(n,200); attach(Auto[Z,]); #
This will be training data of size 200 (for example)
> ss5 = smooth.spline(weight,
mpg, df=5) # Fit the spline using training data only
> attach(Auto);
> Yhat = predict(ss,x=weight)
> names(Yhat) # Notice: prediction consists of two parts
– predictor x and
[1] "x" "y" # predicted response y. We can call them as Yhat$x and Yhat$y.
> mean(( Yhat$y[-Z]
- mpg[-Z] )^2) # Then, compute prediction mean-squared
error on test data
[1] 16.58982 # This is the cross-validation error for a
spline with df=5
R also has a built-in cross-validation tool for smoothing splines. It
uses the leave-one-out method (LOOCV, or jackknife)
> smooth.spline(weight,
mpg, df=5, cv=TRUE)
Smoothing Parameter spar= 1.046238 lambda= 0.01789013 (11 iterations)
Equivalent Degrees of Freedom (Df): 5.000653
Penalized Criterion: 5974.815
PRESS: 17.59263 # This error (PREdiction
Sum of Squares) is computed by LOOCV method
5c. Choosing the optimal degrees of freedom
We’ll fit smoothing splines of various df
and choose the one with the smallest prediction error.
> cv.err =
rep(0,50); Auto.train
= Auto[Z,];
> for (p in 1:50){
+ attach(Auto.train); ss = smooth.spline(weight,
mpg, df=p+0.01) # DF should be > 1
+ attach(Auto); Yhat =
predict(ss, weight)
+ cv.err[p] = mean( (Yhat$y[-Z] - mpg[-Z])^2 ) }
> which.min(cv.err)
[1] 1
# This cross-validation chooses the smoothing spline with the
equivalent of df=1.01
> ss = smooth.spline(weight,
mpg, df=1.01)
> Yhat =
predict(ss)
> plot(weight,mpg);
lines(Yhat, lwd=3)